Anomaly score checks (deprecated)
Anomaly score checks use a machine learning algorithm to automatically detect anomalies in your time-series data.
This check is being deprecated. Soda recommends using the new Metric Monitoring features, rebuilt from the ground up, 70% more accurate and significantly faster.
Use an anomaly score check to automatically discover anomalies in your time-series data.
checks for dim_customer:
- anomaly score for row_count < default✔️ Requires Soda Core Scientific (included in a Soda Agent) ✖️ Supported in Soda Core ✔️ Supported in Soda Library + Soda Cloud ✔️ Supported in Soda Cloud Agreements + Soda Agent ✖️ Available as a no-code check
About anomaly score checks
The anomaly score check is powered by a machine learning algorithm that works with measured values for a metric that occur over time. The algorithm learns the patterns of your data – its trends and seasonality – to identify and flag anomalies in time-series data.
Install Soda Scientific
To use an anomaly score check, you must install Soda Scientific in the same directory or virtual environment in which you installed Soda Library. Soda Scientific is included in Soda Agent deployment. Best practice recommends installing Soda Library and Soda Scientific in a virtual environment to avoid library conflicts, but you can Install Soda Scientific locally if you prefer.
Set up a virtual environment, and install Soda Library in your new virtual environment.
Use the following command to install Soda Scientific.
pip install -i https://pypi.cloud.soda.io soda-scientificList of Soda Scientific dependencies
pandas<2.0.0
wheel
pydantic>=1.8.1,<2.0.0
scipy>=1.8.0
numpy>=1.23.3, <2.0.0
inflection==0.5.1
httpx>=0.18.1,<2.0.0
PyYAML>=5.4.1,<7.0.0
cython>=0.22
prophet>=1.1.0,<2.0.0
Refer to Troubleshoot Soda Scientific installation for help with issues during installation.
Define an anomaly score check
The following example demonstrates how to use the anomaly score for the row_count metric in a check. You can use any numeric, missing, or validity metric in lieu of row_count.
Currently, you can only use
< defaultto define the threshold in an anomaly score check.By default, anomaly score checks yield warn check results, not fails.
You can use any numeric, missing, or validity metric in anomaly score checks. The following example detects anomalies for the average of order_price in an orders dataset.
The following example detects anomalies for the count of missing values in the id column.
Anomaly score check results
Because the anomaly score check requires at least four data points before it can start detecting what counts as an anomalous measurement, the first few scans yield a check result that indicates that Soda does not have enough data.
Though your first instinct may be to run several scans in a row to product the four measurments that the anomaly score needs, the measurements don’t “count” if the frequency of occurrence is too random, or rather, the measurements don't represent enough of a stable frequency.
If, for example, you attempt to run eight back-to-back scans in five minutes, the anomaly score does not register the measurements resulting from those scans as a reliable pattern against which to evaluate an anomaly.
Consider using the Soda library to set up a programmatic scan that produces a check result for an anomaly score check on a regular schedule.
Produce warnings instead of fails
By default, an anomaly score check yields either a pass or fail result; pass if Soda does not detect an anomaly, fail if it does.
If you wish, you can instruct Soda to issue warn check results instead of fails by adding a warn_only configuration, as in the following example.
Reset anomaly history
If you wish, you can reset an anomaly score's history, effectively recalibrating what Soda considers anomalous on a dataset.
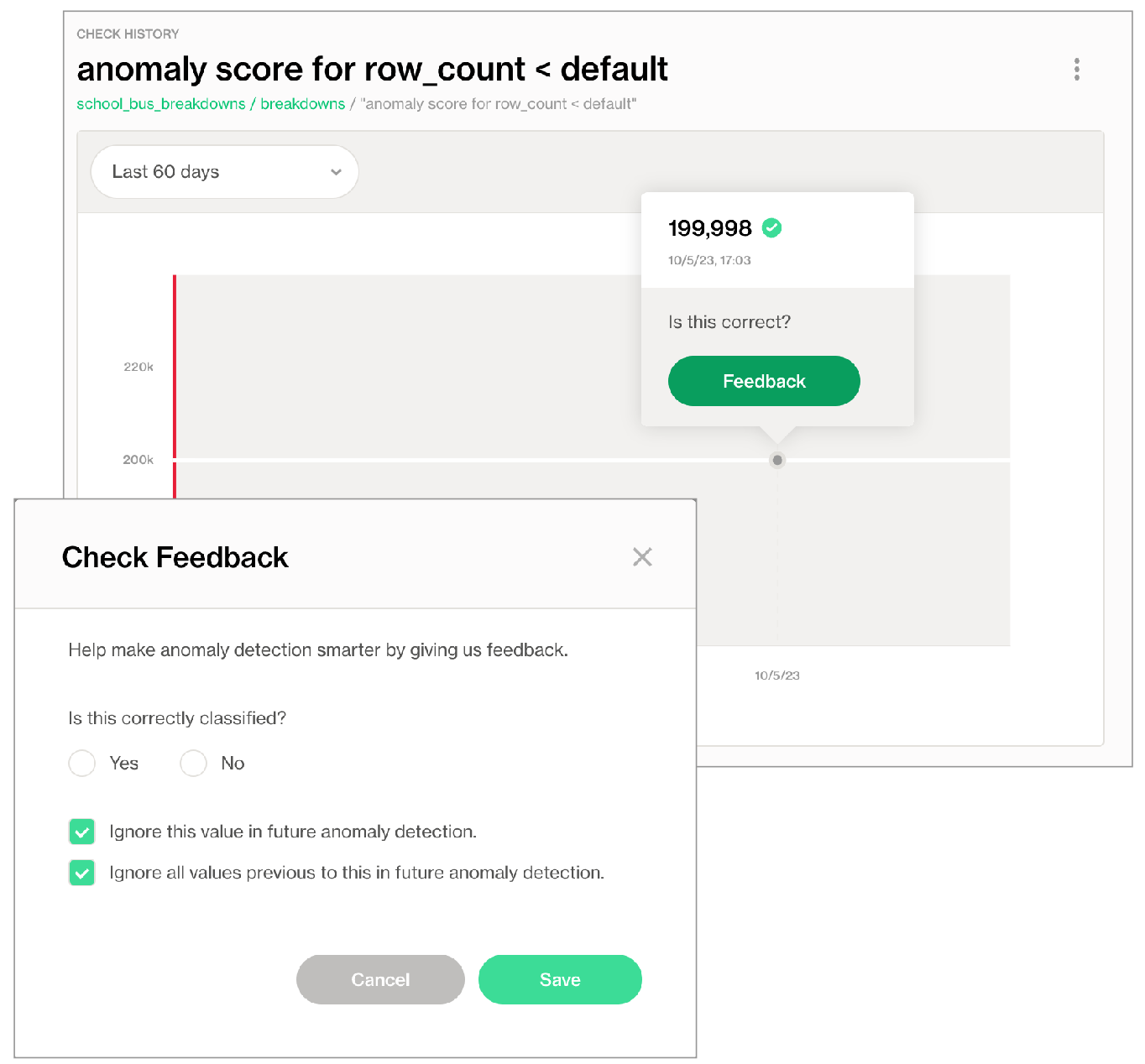
In Soda Cloud, navigate to the Check History page of the anomaly check you wish to reset.
Click to select a node in the graph that represents a measurement, then click Feedback.
In the modal that appears, you can choose to exclude the individual measurement, or all previous data up to that measurement, the latter of which resets the anomaly score's history.
Optional check configurations
✓
Define a name for an anomaly score check.
-
Define alert configurations to specify warn and fail thresholds.
-
✓
Apply an in-check filter to return results for a specific portion of the data in your dataset; see example.
✓
Use quotes when identifying dataset names; see example. Note that the type of quotes you use must match that which your data source uses. For example, BigQuery uses a backtick (`) as a quotation mark.
Use wildcard characters ( % or * ) in values in the check.
-
✓
Use for each to apply anomaly score checks to multiple datasets in one scan; see example.
Example with quotes
Example with for each
Track anomalies and relative changes by group
You can use a group by configuration to detect anomalies by category, and monitor relative changes over time in each category.
✔️ Requires Soda Core Scientific for anomaly check (included in a Soda Agent) ✖️ Supported in Soda Core ✔️ Supported in Soda Library 1.1.27 or greater + Soda Cloud ✔️ Supported in Soda Cloud Agreements + Soda Agent 0.8.57 or greater ✖️ Available as a no-code check
The following example includes three checks grouped by gender.
The first check uses the custom metric
average_childrento collect measurements and gauge them against an absolute threshold of2. Soda Cloud displays the check results grouped by gender.The second check uses the same custom metric to detect anomalous measurements relative to previous measurements. Soda must collect a minimum of four, regular-cadence, measurements to have enough data from which to gauge an anomolous measurement. Until it has enough measurements, Soda returns a check result of
[NOT EVALUATED]. Soda Cloud displays any detected anomalies grouped by gender.The third check uses the same custom metric to detect changes over time in the calculated average measurement, and gauge the measurement against a threshold of
between -5 and 5relative to the previously-recorded measurement. See Change-over-time thresholds for supported syntax variations for change-over-time checks. Soda Cloud displays any detected changes grouped by gender.

Troubleshoot Soda Scientific installation
While installing Soda Scientific works on Linux, you may encounter issues if you install Soda Scientific on Mac OS (particularly, machines with the M1 ARM-based processor) or any other operating system. If that is the case, consider using one of the following alternative installation procedures.
Need help? Ask the team in the Soda community on Slack.
Install Soda Scientific Locally
Set up a virtual environment, and install Soda Library in your new virtual environment.
Use the following command to install Soda Scientific.
List of Soda Scientific dependencies
pandas<2.0.0
wheel
pydantic>=1.8.1,<2.0.0
scipy>=1.8.0
numpy>=1.23.3, <2.0.0
inflection==0.5.1
httpx>=0.18.1,<2.0.0
PyYAML>=5.4.1,<7.0.0
cython>=0.22
prophet>=1.1.0,<2.0.0
Use Docker to run Soda Library
Use Soda’s Docker image in which Soda Scientific is pre-installed. You need Soda Scientific to be able to use SodaCL distribution checks or anomaly detection checks.
If you have not already done so, install Docker in your local environment.
From Terminal, run the following command to pull Soda Library’s official Docker image; adjust the version to reflect the most recent release.
Verify the pull by running the following command.
Output:
When you run the Docker image on a non-Linux/amd64 platform, you may see the following warning from Docker, which you can ignore.
When you are ready to run a Soda scan, use the following command to run the scan via the docker image. Replace the placeholder values with your own file paths and names.
Optionally, you can specify the version of Soda Library to use to execute the scan. This may be useful when you do not wish to use the latest released version of Soda Library to run your scans. The example scan command below specifies Soda Library version 1.0.0.
What does the scan command do?
docker runensures that the docker engine runs a specific image.-vmounts your SodaCL files into the container. In other words, it makes the configuration.yml and checks.yml files in your local environment available to the docker container. The command example maps your local directory to/sodaclinside of the docker container.sodadata/soda-libraryrefers to the image thatdocker runmust use.scaninstructs Soda Library to execute a scan of your data.-dindicates the name of the data source to scan.-cspecifies the filepath and name of the configuration YAML file.
Error: Mounts denied
If you encounter the following error, follow the procedure below.
You need to give Docker permission to acccess your configuration.yml and checks.yml files in your environment. To do so:
Access your Docker Dashboard, then select Preferences (gear symbol).
Select Resources, then follow the Docker instructions to add your Soda project directory – the one you use to store your configuration.yml and checks.yml files – to the list of directories that can be bind-mounted into Docker containers.
Click Apply & Restart, then repeat steps 2 - 4 above.
Error: Configuration path does not exist
If you encounter the following error, double check the syntax of the scan command in step 4 above.
Be sure to prepend
/sodacl/to both the congifuration.yml filepath and the checks.yml filepath.Be sure to mount your files into the container by including the
-voption. For example,-v /Users/MyName/soda_project:/sodacl.
Troubleshoot Soda Scientific installation in a virtual env
If you have defined an anomaly detection check and you use an M1 MacOS machine, you may get aLibrary not loaded: @rpath/libtbb.dylib error. This is a known issue in the MacOS community and is caused by issues during the installation of the prophet library. There currently are no official workarounds or releases to fix the problem, but the following adjustments may address the issue.
Install
soda-scientificas per the local environment installation instructions and activate the virtual environment.Use the following command to navigate to the directory in which the
stan_modelof theprophetpackage is installed in your virtual environment.For example, if you have created a python virtual environment in a
/venvsdirectory in your home directory and you use Python 3.9, you would use the following command.Use the
lscommand to determine the version number ofcmndstanthatprophetinstalled. Thecmndstandirectory name includes the version number.Add the
rpathof thetbblibrary to yourprophetinstallation using the following command.With
cmdstanversion2.26.1, you would use the following command.
List of comparison symbols and phrases
Go further
Reference tips and best practices for SodaCL.
Need help? Join the Soda community on Slack.
Last updated
Was this helpful?
